What are some current nuclease resistance modifications that are being used?
Oligonucleotides are susceptible to enzymatic degradation by exonucleases and endonucleases, particularly RNA, which is prone to both chemical instability and rapid breakdown. These challenges can significantly shorten their half-life and limit their effectiveness. To counteract these issues, various chemical modifications can be incorporated to improve stability, enhance nuclease resistance, and prolong oligo lifespan. These strategies are crucial for ensuring consistent performance in laboratory studies, diagnostics, and therapeutic settings.
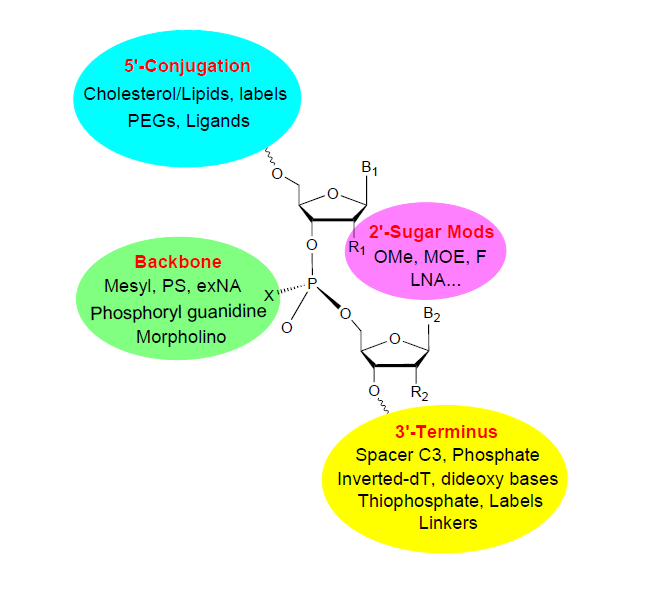
Backbone:
- Phosphorothioate (PS): A phosphodiester modification where a non-bridging oxygen atom is replaced with sulfur, resulting in the formation of stereoisomers (Rp and Sp). The Sp configuration at the 3′ end enhances resistance to 3′ exonucleases, which preferentially recognize the Rp configuration. However, PS modifications can introduce toxicity at higher concentrations, requiring careful design and application [1, 2].
- Mesyl Phosphoramidate (MsPA): A methanesulfonyl group replaces the natural phosphodiester group, providing exceptional nuclease resistance while preserving the oligonucleotide’s hybridization properties. This modification is highly effective in improving oligonucleotide stability for various applications [3].
- Morpholinos: These oligonucleotide analogs substitute the ribose-phosphate backbone with a morpholine ring, a non-ionic structure that is highly resistant to enzymatic degradation. Morpholinos are widely employed in gene knockdown experiments due to their stability and efficient target binding in biological systems [4].
- Extended Nucleic Acid (exNA): A recent study highlights a novel modification to the siRNA backbone involving the insertion of a methylene group between the 5′C and 5′OH of a nucleoside. This modification, when combined with phosphorothioate (PS) linkages, provides remarkable resistance to nuclease degradation, achieving a 32-fold increase in protection against both 3′ and 5′ exonucleases. The enhanced stability is attributed to the exNA backbone’s ability to sterically hinder nuclease activity by inhibiting the initial binding [5].
2′ Sugar:
These 2’ modifications below are often used in conjunction with other modifications like PS backbone and nucleic acids for ASOs, splice switching oligos (SSOs), and siRNA.
- 2′-O-Methyl (2′-OMe): This modification adds a methyl group to the 2′-hydroxyl of the ribose, enhancing thermal stability by forming homoduplexes with higher melting temperatures (Tm) compared to RNA:RNA and DNA:DNA duplexes. The stabilization effect is most pronounced when 2′-OMe is incorporated uniformly, as it promotes a consistent sugar pucker conformation across the oligo rather than through point substitutions. Additionally, 2′-OMe modifications improve nuclease resistance, likely due to the enhanced stability of the secondary structure or duplex formation [6]
- 2′-O-methoxyethyl (2′-MOE): Similar to 2′-OMe, this modification replaces the natural 2′-hydroxyl group with a methoxyethyl group. The larger methoxyethyl moiety offers steric hindrance that protects against nuclease degradation while also promoting a stable duplex formation. Studies show the methoxyethyl group projects into the minor groove of the duplex, enforcing a C3′-endo sugar pucker that enhances affinity. Notably, tethering the 2′-MOE group to the 4′ position of the furanose ring can yield a constrained 2′,4′-MOE structure for even greater stability. [7].
- 2′-Fluoro (2′-F): Incorporating 2′-Fluoro groups alongside phosphorothioate (PS) linkages uniformly across the oligo improves thermal stability and melting temperature. RNA duplexes naturally favor the 3′-endo sugar conformation, and the 2′-Fluoro modification stabilizes this structure, enhancing the thermodynamic stability of RNA-antisense duplexes. By replacing the 2′-hydroxyl group with fluorine, the oligo becomes resistant to nucleases targeting this hydroxyl group [8].
- Locked Nucleic Acids (LNA): LNA modifications introduce a methylene bridge that locks the ribose sugar in a rigid conformation, enhancing hybridization affinity and resistance to nucleases. Due to their strong thermodynamic properties, LNAs are often combined with other linkages to offset low binding affinity or further stabilize duplexes [2,9].
3′ End Modification:
- Phosphorylation: Adding a phosphate group at the 3′ end is a widely utilized strategy to enhance nuclease resistance. This modification blocks the access of exonucleases that rely on a free 3′ hydroxyl group to initiate digestion, thereby acting as an effective barrier.
- 3’ Inverted dT: Incorporating an inverted thymidine at the 3′ end creates a 3′-3′ linkage, where the 3′ carbon of the oligo connects to the 3′ carbon of the inverted thymidine instead of the standard 5′ phosphodiester bond. This unusual configuration disrupts the active site of exonucleases, preventing degradation. It is a popular choice for aptamers and other functional oligos [10].
- Threofuranosyl nucleic acid (TNA): TNA demonstrates exceptional nuclease resistance, with studies showing its stability after incubation with snake venom phosphodiesterase (SVPDE) at 37°C for seven days. This resistance is attributed to the replacement of the natural sugar backbone with threose, an artificial polymer less recognizable by nucleases. TNA also forms stable antiparallel Watson-Crick duplexes with complementary RNA and DNA strands, leveraging its A-like helical geometry to shield the oligos from enzymatic digestion [11].
- Methyl phosphonate (MP): Methyl phosphonate linkages exhibit total resistance to spleen phosphodiesterase activity. A study reveals that consecutive MP linkages are necessary to prevent exonucleases like SVPDE from “jumping” single modifications and resuming activity. The neutral charge of MP linkages likely slows enzyme dissociation from the active site, resulting in reduced hydrolysis rates. Oligos with 3′ MP linkages show superior resistance compared to those modified at both the 5′ and 3′ ends [2].
5′ End Modification:
- Dialkyl sulfide (s) linkage: Kawai et al. developed 5′-SH functionalized thymidine dinucleotides with dialkyl sulfide (S) linkages (TsT) and tested them against various nucleases, including SVPDE, CSPDE, nuclease S1, and P1, at 37°C for 48 hours. None of these enzymes were able to hydrolyze the sulfide linkage, likely due to steric hindrance from the sulfur and alkyl groups. However, SVPDE showed the ability to bypass the modification, degrading a 6-mer d(TsTTsTTsT) to dimers (TsT). To achieve effective protection, consecutive modified linkages are recommended [2].
- Thiophosphate (SP) linkage: In a 5′-SP modified oligonucleotide (ODN), one bridging oxygen atom is substituted with a sulfur atom. This modification enhances nuclease resistance, potentially due to changes in the binding length and altered electrostatic interactions within the enzyme’s active site. In gapmers, 5′-SP linkages exhibit the highest relative stability when positioned at the 5′ end of the gap [2].
- Triazole (TR)-LNA linkage: While triazole alone tends to reduce stability due to its rigidity, combining it with locked nucleic acid (LNA) enhances duplex stability. Incorporating three triazole modifications with LNA at the 5′ end improves siRNA duplex resistance against the 5′ exonuclease XRN1 [2].
- Boronate (bn) Linkage Boronate modifications involve replacing the natural phosphodiester linkage with a boronate ester (Bn) at the 5′ end, creating a reversible covalent bond with cis-diol groups on the sugar backbone. This shields the cleavage site from nucleases and increases resistance to SVPDE, exonuclease III, and CSPDE. Additionally, 5′ boronic acid can inhibit RNase H activity, while oxidation of the boronate group can restore enzymatic activity [2].
Conjugation:
- Cholesterol or Lipid: Cholesterol conjugation enhances cellular uptake by facilitating passage across plasma membranes. This modification improves delivery efficiency and provides protection from enzymatic degradation, ensuring greater oligonucleotide stability in biological environments [12].
- PEGylation: A study demonstrated that PEGylation of complementary RNA oligos at the 5′ end significantly stabilized mRNA, increasing its nuclease resistance by 15-fold during serum incubation. Despite adding up to 15 PEG strands, the mRNA size remained compact. The PEG strands are hypothesized to form a steric and electrostatic shield around the mRNA, reducing enzymatic degradation and enhancing stability [13].
References:
- Iwamoto, N., Butler, D., Svrzikapa, N. et al. Control of phosphorothioate stereochemistry substantially increases the efficacy of antisense oligonucleotides. Nat Biotechnol 35, 845–851 (2017). https://doi.org/10.1038/nbt.3948
- Clavé G, Reverte M, Vasseur JJ, Smietana M. Modified internucleoside linkages for nuclease-resistant oligonucleotides. RSC Chem Biol. 2020 Dec 8;2(1):94-150. doi: 10.1039/d0cb00136h.
- Patutina OA, Gaponova Miroshnichenko SK, Sen’kova AV, Savin IA, Gladkikh DV, Burakova EA, Fokina AA, Maslov MA, Shmendel’ EV, Wood MJA, Vlassov VV, Altman S, Stetsenko DA, Zenkova MA. Mesyl phosphoramidate backbone modified antisense oligonucleotides targeting miR-21 with enhanced in vivo therapeutic potency. Proc Natl Acad Sci U S A. 2020 Dec 22;117(51):32370-32379. doi: 10.1073/pnas.2016158117.
- Hudziak, Barofsky, Barofsky, Weller, Huang, Weller (1996). Resistance of Morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucleic Acid Drug Dev. 6 , 267.
- Yamada, K., Hariharan, V.N., Caiazzi, J. et al. Enhancing siRNA efficacy in vivo with extended nucleic acid backbones. Nat Biotechnol (2024). https://doi.org/10.1038/s41587-024-02336-7.
- Cummins LL, Owens SR, Risen LM, Lesnik EA, Freier SM, McGee D, Guinosso CJ, Cook PD. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995 Jun 11;23(11):2019-24. doi: 10.1093/nar/23.11.2019.
- Pallan PS, Allerson CR, Berdeja A, Seth PP, Swayze EE, Prakash TP, Egli M. Structure and nuclease resistance of 2′,4′-constrained 2′-O-methoxyethyl (cMOE) and 2′-O-ethyl (cEt) modified DNAs. Chem Commun (Camb). 2012 Aug 25;48(66):8195-7. doi: 10.1039/c2cc32286b.
- Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J Med Chem. 1993 Apr 2;36(7):831-41. doi: 10.1021/jm00059a007.
- Bao T. Le, Abbie M. Adams, Susan Fletcher, Stephen D. Wilton, Rakesh N. Veedu. Rational Design of Short Locked Nucleic Acid-Modified 2′-O-Methyl Antisense Oligonucleotides for Efficient Exon-Skipping In Vitro, Molecular Therapy – Nucleic Acids. Volume 9. 2017. Pages 155-161. ISSN 2162-2531. https://doi.org/10.1016/j.omtn.2017.09.002.
- Ni S, Yao H, Wang L, Lu J, Jiang F, Lu A, Zhang G. Chemical Modifications of Nucleic Acid Aptamers for Therapeutic Purposes. International Journal of Molecular Sciences. 2017; 18(8):1683. https://doi.org/10.3390/ijms18081683.
- Culbertson MC, Temburnikar KW, Sau SP, Liao JY, Bala S, Chaput JC. Evaluating TNA stability under simulated physiological conditions. Bioorg Med Chem Lett. 2016 May 15;26 (10):2418-2421. doi: 10.1016/j.bmcl.2016.03.118.
- Obermann Hannah-Lena , Lederbogen Ines I. , Steele Jenny , Dorna Jens , Sander Leif Erik , Engelhardt Konrad , Bakowsky Udo , Kaufmann Andreas , Bauer Stefan. RNA-Cholesterol Nanoparticles Function as Potent Immune Activators via TLR7 and TLR8. Frontiers in Immunology. Volume 12. 2022. https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2021.658895.
- Yoshinaga, N.; Naito, M.; Tachihara, Y.; Boonstra, E.; Osada, K.; Cabral, H.; Uchida, S. PEGylation of mRNA by Hybridization of Complementary PEG-RNA Oligonucleotides Stabilizes mRNA without Using Cationic Materials. Pharmaceutics 2021, 13, 800. https://doi.org/10.3390/pharmaceutics13060800
