What is Spatial Transcriptomics?
Spatial transcriptomics is a cutting-edge technology that allows researchers to study gene expression profiles of cells while maintaining their location within a tissue. This method has become popular in fields like oncology, neurobiology, and developmental biology due to its ability to map gene expression at single-cell resolution. By preserving tissue architecture, it provides deep insights into how cells are organized and function. Commonly used methods include spatial barcoding and in situ hybridization.
Why MERFISH is being used in Spatial Transcriptomics?
Multiplexed Error-Robust Fluorescence In Situ Hybridization (MERFISH) is a powerful tool in spatial transcriptomics. This advanced single-molecule imaging technique enables the visualization and quantification of thousands of RNA transcripts with high spatial resolution and error robustness, making it ideal for tissue mapping, single-cell gene expression studies, and cancer research.
MERFISH has key advantages, such as retaining spatial context in cells’ natural environments. Its high throughput and single-molecule sensitivity allow measurements of thousands of genes in one experiment, delivering highly precise gene expression data. A recent study using branched DNA (bDNA) amplification improved MERFISH’s performance, achieving nearly 100% detection efficiency. This method increases signal brightness and reduces the number of probes needed for each RNA species.
What kind of probes are used in MERFISH?
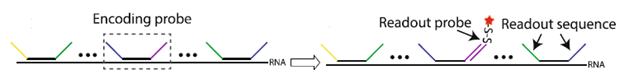
MERFISH relies on several types of probes for multiplexed RNA detection. The primary (encoding) probes bind to different regions of the RNA molecule, while the secondary (readout) probes carry fluorescent tags and hybridize with the encoding probes during imaging.
Each encoding probe consists of a primer region (for PCR amplification), two readout sequences (each assigned a binary barcode for the RNA they bind to), and a targeting sequence (~30 nucleotides) (Refer to Figure 1). Multiple rounds of hybridization and imaging identify the readout sequences, and the binding of the readout probes determines the bits that are linked to a specific binary code. Multiple encoding probes can target different regions of the same RNA, increasing the signal from each RNA copy [2].
Can Synoligo make MERFISH probes?
Yes! Synoligo specializes in synthesizing MERFISH encoding and readout probes with fast turnaround times. Here is the general format of a readout probe, where the fluorophore can be any of your choosing (FAM™, Cy™3/5, Alexa™ or ATTO™ dyes):
5’-fluorophore -C6-S-S-C6 linker (cleavable)-NNNNNN (DNA sequence)-3’
When designing probes, it’s important to maintain consistent GC content and melting temperature (Tm) across target regions and to avoid homology with other RNAs to ensure probe specificity.
References:
- Xia, C., Babcock, H.P., Moffitt, J.R. et al. Multiplexed detection of RNA using MERFISH and branched DNA amplification. Sci Rep 9, 7721 (2019). https://doi.org/10.1038/s41598-019-43943-8
- Moffitt, J. R., & Zhuang, X. (2016). RNA Imaging with Multiplexed Error-Robust Fluorescence In Situ Hybridization (MERFISH). Methods in enzymology, 572, 1–49. https://doi.org/10.1016/bs.mie.2016.03.020
