A recent paper by Wave Life Sciences1 describes a new application of isouridine (N3U) for improving the efficiency of ADAR-assisted editing of mRNA in a disease-relevant target.
Background
ADARs (Adenosine Deaminase Acting on RNA) are endogenous enzymes that are found in all vertebrates. In humans ADAR1 and ADAR2 bind dsRNA and perform A-to-I edits at the target site and are found in all tissues of the body, whereas ADAR3 is localized to the brain and lacks the catalytic site. ADAR1 and ADAR2 are essential proteins and knockouts are lethal with ADAR1 suppressing aberrant immune responses against self-dsRNA by disrupting the RNA duplex by introducing I-U mismatches and bulges.2 The reaction is shown below, showing the changes in the hydrogen bond donor and acceptors.
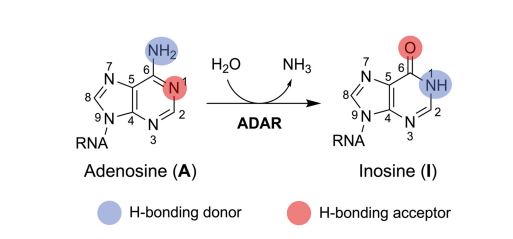
Reprinted from © 2024 Mendoza and Beal; Published by Cold Spring Harbor Laboratory Press for the RNA Society under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/
Therapeutic Potential of Isouridine (N3U) in ADAR Editing
The potential of using ADARs for therapeutic purposes has not gone unnoticed by the research community. ADAR1 and ADAR2 are ubiquitous and found in all the cells in the body and present a clear opportunity to treat diseases with G-to-A point mutations. This is because the A-to-I edited base in the mRNA transcript is read by translation machinery as a G, restoring the function of the expressed protein. By introducing antisense RNA oligos that target the region of the G-to-A point mutation, endogenous ADARs can be recruited to edit the dysfunctional mRNA transcript. This strategy has the advantage over genome editing in that the issues of permanent, off-target editing are avoided.
The importance of isouridine – aka (N3U)
The difficulty with using ADARs to target G-to-A point mutations is the strong sequence dependence on the 5’ and 3’ bases that flank the target. ADAR1 and ADAR2 have the following sequence preferences for the adjacent 5’ and 3’ bases: 5’X A Y3’
ADAR1 X: U>A>C>G Y: G>C~A>U
ADAR2 X: U>A>C>G Y: G>C>A~U
So, a sequence such as 5’-..NNN GAU NNN..-3’ will be edited with poor efficiency. These preferences are not due to sequence recognition or specificity but rather their effect upon the rate of the adenosine flipping into the active site of the ADAR3.
The second critical factor in editing efficiency is the “orphan base” which is the base in the ASO that sits opposite to the A in the targeted sequence. The preferred orphan base that leads to the highest editing efficiency is C. The C base forms hydrogen bonds with a conserved glutamate residue that is found in both ADAR1 and ADAR2 which facilitates the flipping of the A into the active site. A and G both clash with the glutamate and U, while it can form a hydrogen bond to the glutamate, has to do so at the energetic expense of breaking its existing hydrogen bonds to the A.
The scientists at Wave Life Sciences used molecular modeling to find an analog that would
- form hydrogen bonds to the conserved glutamate residue
- not form hydrogen bonds to A
The analog that best fit these requirements was the isouridine (N3U).
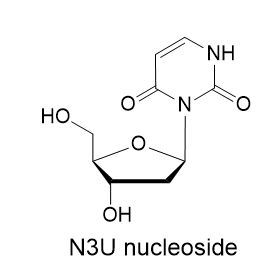
Molecular modeling suggested the N3U analog in the orphan position of the dsRNA would form hydrogen bonds to the conserved glutamate in the ADAR1, however it could only form a single hydrogen bond with the A targeted for deamination. Therefore, the N3U in the orphan position should increase the rate of the flipping of A into the ADAR active site and improve editing efficiency. The N3U analog also had the advantage of being relatively simple to synthesize and was well-behaved as a phosphoramidite.
This analog was tested in Wave Life Sciences “AIMer” platform for ADAR editing, looking at combinations of 2’-OMe, 2’-F, 2’-deoxy sugars as well as backbone modifications such as phosphorothioate (PS), phosphoryl guanidine, phosphodiester, with some stereospecific Rp and Sp linkages within the sequence. Incorporating the N3U in the orphan position of the AIMer led to broadly consistent improvements in A-to-I editing efficiency in the target mRNA. In one AIMmer, the editing efficiency increased from 2% using C in the orphan position to a remarkable 82% with N3U for the identical sequence.
Finally, the authors demonstrated the efficacy of N3U in a disease-relevant target, SERPINA1 that contains a G-to-A point mutation that leads to α1-antitrypsin deficiency. The sugar and backbone modifications were explored with the most promising sequence, SERPINA1-989, having 2’-F and 2’-OMe sugar modifications distributed throughout the sequence along with Sp PS and both Rp and Sp phorphoryl guanidine backbone modifications, gave the best overall performance. In addition, for this work a large number of sugar variants of N3U itself were tested including 2’-OMe-N3U, 2’-fluoro-N3U, 2’-FANA-N3U, 2’-MOE-N3U and others, however, the 2’-deoxy-N3U actually performed better overall.
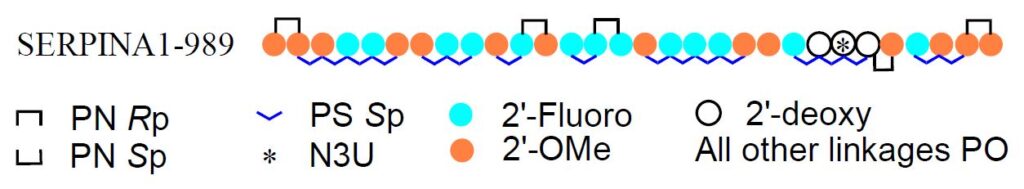
Take home message
The A-to-I editing efficiency significantly and consistently increased – and sometimes dramatically – when using 2’-deoxy-N3U in the orphan position of the AIMer compared to C. These improvements were consistent in a large variety of AIMer sequences and against different mRNA targets. Editing efficiency could further be increased by increasing nuclease resistance of the AIMer by using 2’-OMe and 2’-Fluoro RNA bases spread out through the sequence in addition to phosphorothioate and phosphoryl guanidine backbone linkages.
Synoligo has the experience and expertise to deliver N3U/2’-F/2’-OMe/2’-MOE-modified oligos with stereospecific Rp and Sp backbone modifications including phosphorothioate, phosphoryl guanidine, and other phosphoramidate (PN) linkages. Contact our team to discuss how we can help you with your project.
References:
- Lu, Genliang, et al. “Rational design of base, sugar and backbone modifications improves ADAR-mediated RNA editing.” Nucleic Acids Research 52.17 (2024): 10068-10084.
- Yang, Yuxi, Shunpei Okada, and Masayuki Sakurai. “Adenosine-to-inosine RNA editing in neurological development and disease.” RNA biology 18.7 (2021): 999-1013.
- Kuttan, Ashani, and Brenda L. Bass. “Mechanistic insights into editing-site specificity of ADARs.” Proceedings of the National Academy of Sciences 109.48 (2012): E3295-E3304.
